# R console
install.packages(c("biglm", "dplyr", "ggplot2", "readr", "targets", "tidyr"))Appendix C — Target Markdown
C.1 Superseded
Target Markdown is superseded. Please instead use the tar_tangle() function from the tarchetypes package.
C.2 About
Target Markdown, available in targets > 0.6.0, is knitr-based interface for reproducible analysis pipelines.1 With Target Markdown, you can define a fully scalable pipeline from within one or more Quarto or R Markdown reports or projects (even spreading a single pipeline over multiple source documents). You get the best of both worlds: the human readable narrative of literate programming, and the sophisticated caching and dependency management systems of targets.
C.3 Access
This chapter’s example Target Markdown document is itself a tutorial and a simplified version of the chapter. There are two convenient ways to access the file:
- The
use_targets()function. - The RStudio R Markdown template system.
For (2), in the RStudio IDE, select a new Quarto or R Markdown document in the New File dropdown menu in the upper left-hand corner of the window.
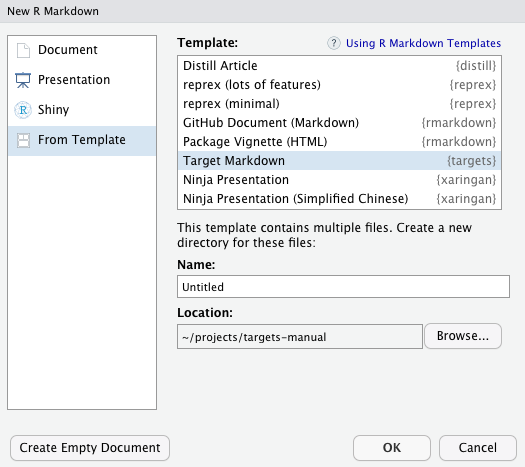
Then, select the Target Markdown template and click OK to open a copy of the report for editing.
C.4 Purpose
Target Markdown has two primary objectives:
- Interactively explore, prototype, and test the components of a
targetspipeline using the Quarto notebook interface or the R Markdown notebook interface. - Set up a
targetspipeline using convenient Markdown-like code chunks.
Target Markdown supports a special {targets} language engine with an interactive mode for (1) and a non-interactive mode for (2). By default, the mode is interactive in the notebook interface and non-interactive when you knit/render the whole document.2. You can set the mode using the tar_interactive chunk option.
C.5 Example
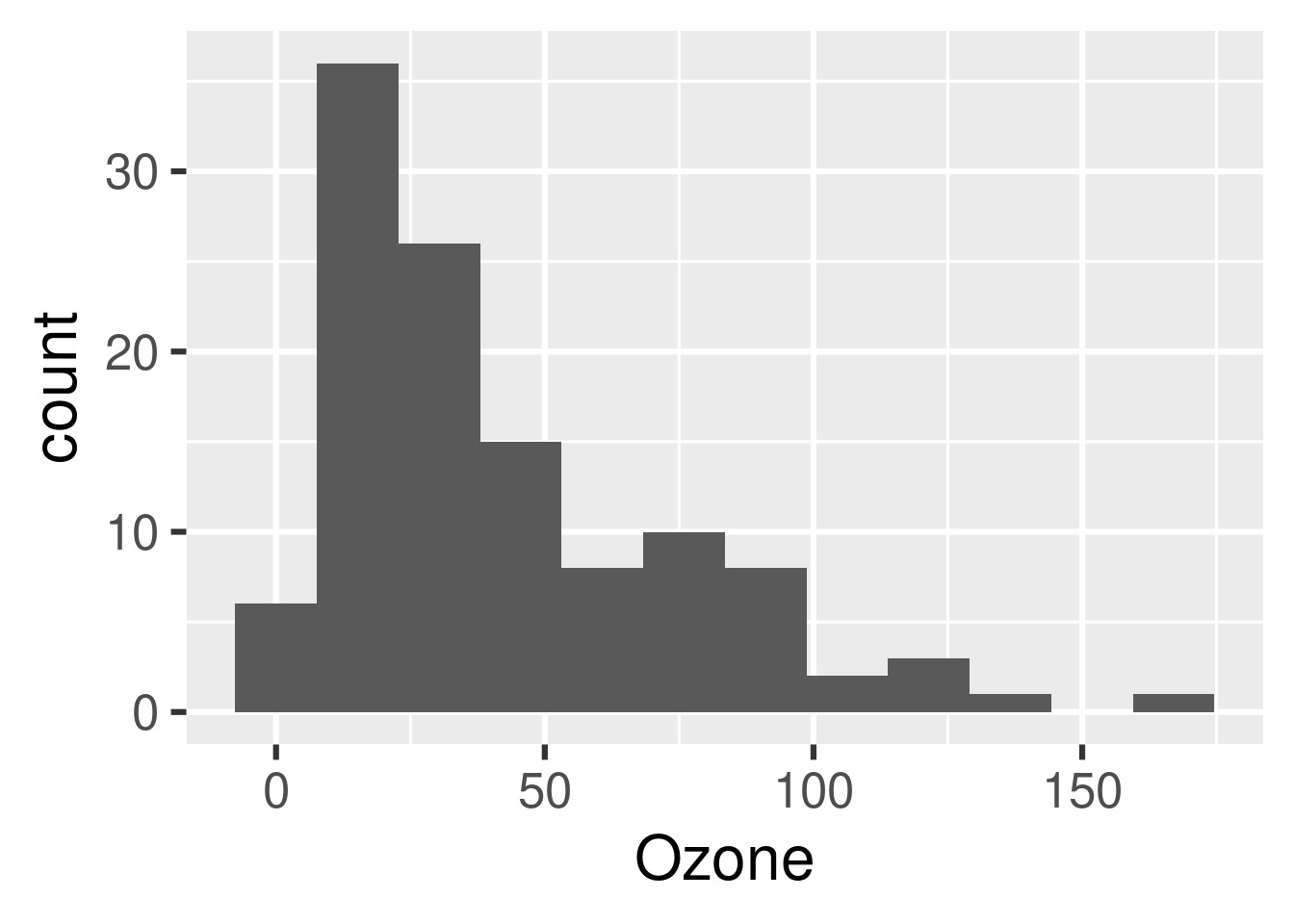
The following example is based on the minimal targets project at https://github.com/wlandau/targets-minimal/. We process the base airquality dataset, fit a model, and display a histogram of ozone concentration.
C.6 Required packages
This example requires several R packages, and targets must be version 0.6.0 or above.
C.7 Setup
First, load targets to activate the specialized knitr engine for Target Markdown.
```{r}
library(targets)
library(tarchetypes)
```Non-interactive Target Markdown writes scripts to a special _targets_r/ directory to define individual targets and global objects. In order to keep your target definitions up to date, it is recommended to remove _targets_r/ at the beginning of the R Markdown document(s) in order to clear out superfluous targets and globals from a previous version. tar_unscript() is a convenient way to do this.
```{r}
tar_unscript()
```C.8 Globals
As usual, your targets depend on custom functions, global objects, and tar_option_set() options you define before the pipeline begins. Define these globals using the {targets} engine with tar_globals = TRUE chunk option.
```{targets some-globals, tar_globals = TRUE, tar_interactive = TRUE}
options(tidyverse.quiet = TRUE)
tar_option_set(packages = c("biglm", "dplyr", "ggplot2", "readr", "tidyr"))
create_plot <- function(data) {
ggplot(data) +
geom_histogram(aes(x = Ozone), bins = 12) +
theme_gray(24)
}
```In interactive mode, the chunk simply runs the R code in the tar_option_get("envir") environment (usually the global environment) and displays a message:
#> Run code and assign objects to the environment.Here is the same chunk in non-interactive mode. Normally, there is no need to duplicate chunks like this, but we do so here in order to demonstrate both modes.
```{targets chunk-name, tar_globals = TRUE, tar_interactive = FALSE}
options(tidyverse.quiet = TRUE)
tar_option_set(packages = c("biglm", "dplyr", "ggplot2", "readr", "tidyr"))
create_plot <- function(data) {
ggplot(data) +
geom_histogram(aes(x = Ozone), bins = 12) +
theme_gray(24)
}
```In non-interactive mode, the chunk establishes a common _targets.R file and writes the R code to a script in _targets_r/globals/, and displays an informative message:3
#> Establish _targets.R and _targets_r/globals/chunk-name.R.It is good practice to assign explicit chunk labels or set the tar_name chunk option on a chunk-by-chunk basis. Each chunk writes code to a script path that depends on the name, and all script paths need to be unique.4
C.9 Target definitions
To define targets of the pipeline, use the {targets} language engine with the tar_globals chunk option equal FALSE or NULL (default). The return value of the chunk must be a target definition object or a list of target definition objects, created by tar_target() or a similar function.
Below, we define a target to establish the air quality dataset in the pipeline.
```{targets raw-data, tar_interactive = TRUE}
tar_target(raw_data, airquality)
```If you run this chunk in interactive mode, the target’s R command runs, the engine tests if the output can be saved and loaded from disk correctly, and then the return value gets assigned to the tar_option_get("envir") environment (usually the global environment).
#> Run targets and assign them to the environment.In the process, some temporary files are created and destroyed, but your local file space will remain untouched (barring any custom side effects in your custom code).
After you run a target in interactive mode, the return value is available in memory, and you can write an ordinary R code chunk to read it.
```{r}
head(raw_data)
```The output is the same as what tar_read(raw_data) would show after a serious pipeline run.
head(raw_data)
#> Ozone Solar.R Wind Temp Month Day
#> 1 41 190 7.4 67 5 1
#> 2 36 118 8.0 72 5 2
#> 3 12 149 12.6 74 5 3
#> 4 18 313 11.5 62 5 4
#> 5 NA NA 14.3 56 5 5
#> 6 28 NA 14.9 66 5 6For demonstration purposes, here is the raw_data target code chunk in non-interactive mode.
```{targets chunk-name-with-target, tar_interactive = FALSE}
tar_target(raw_data, airquality)
```In non-interactive mode, the {targets} engine does not actually run any targets. Instead, it establishes a common _targets.R and writes the code to a script in _targets_r/targets/.
#> Establish _targets.R and _targets_r/targets/chunk-name-with-target.R.Next, we define more targets to process the raw data and plot a histogram. Only the returned value of the chunk code actually becomes part of the pipeline, so if you define multiple targets in a single chunk, be sure to wrap them all in a list.
```{targets downstream-targets}
list(
tar_target(data, raw_data %>% filter(!is.na(Ozone))),
tar_target(hist, create_plot(data))
)
```In non-interactive mode, the whole target list gets written to a single script.
#> Establish _targets.R and _targets_r/targets/downstream-targets.R.Lastly, we define a target to fit a model to the data. For simple targets like this one, we can use convenient shorthand to convert the code in a chunk into a valid target. Simply set the tar_simple chunk option to TRUE.
```{targets fit, tar_simple = TRUE}
analysis_data <- data
biglm(Ozone ~ Wind + Temp, analysis_data)
```When the chunk is preprocessed, chunk label (or the tar_name chunk option if you set it) becomes the target name, and the chunk code becomes the target command. All other arguments of tar_target() remain at their default values (configurable with tar_option_set() in a tar_globals = TRUE chunk). The output in the rendered R Markdown document reflects this preprocessing.
tar_target(fit, {
biglm(Ozone ~ Wind + Temp, data)
})
#> Define target fit from chunk code.
#> Establish _targets.R and _targets_r/targets/fit.R.C.9.1 Pipeline
If you ran all the {targets} chunks in non-interactive mode (i.e. pipeline construction mode), then the target script file and helper scripts should all be established, and you are ready to run the pipeline in with tar_make() in an ordinary {r} code chunk. This time, the output is written to persistent storage at the project root.
```{r}
tar_make()
```#> • start target raw_data
#> • built target raw_data [0.585 seconds]
#> • start target data
#> • built target data [0.009 seconds]
#> • start target fit
#> • built target fit [0.003 seconds]
#> • start target hist
#> • built target hist [0.014 seconds]
#> • end pipeline [0.765 seconds]C.9.2 Output
You can retrieve results from the _targets/ data store using tar_read() or tar_load().
```{r}
library(biglm)
tar_read(fit)
```#> Large data regression model: biglm(Ozone ~ Wind + Temp, data)
#> Sample size = 116```{r}
tar_read(hist)
```
The targets dependency graph helps your readers understand the steps of your pipeline at a high level.
```{r}
tar_visnetwork()
```At this point, you can go back and run {targets} chunks in interactive mode without interfering with the code or data of the non-interactive pipeline.
C.10 Conditioning on interactive mode
targets version 0.6.0.9001 and above supports the tar_interactive() function, which suppresses code unless Target Markdown interactive mode is turned on. Similarly, tar_noninteractive() suppresses code in interactive mode, and tar_toggle() selects alternative pieces of code based on the current mode.
C.11 tar_interactive()
tar_interactive() is useful for dynamic branching. If a dynamic target branches over a target from a different chunk, this ordinarily breaks interactive mode.
```{targets condition, tar_interactive = TRUE}
tar_target(y, x ^ 2, pattern = map(x))
```#> Run targets and assign them to the environment.
#> Error:
#> ! Target y tried to branch over x, which is illegal...However, with tar_interactive(), you can define a version of x just for testing and prototyping in interactive mode. The chunk below fixes interactive mode without changing the pipeline in non-interactive mode.
```{targets condition-fixed, tar_interactive = TRUE}
list(
tar_interactive(tar_target(x, seq_len(2))),
tar_target(y, x ^ 2, pattern = map(x))
)
```#> Run targets and assign them to the environment.C.12 tar_toggle()
tar_toggle() is useful for scaling up and down the amount of work based on the current mode. Interactive mode should finish quickly for prototyping and testing, and non-interactive mode should take on the full level work required for a serious pipeline. Below, tar_toggle() seamlessly scales up and down the number of simulations repetitions in the example target from https://wlandau.github.io/rmedicine2021-pipeline/#target-definitions. To learn more about stantargets, visit https://docs.ropensci.org/stantargets/.
```{targets bayesian-model-validation, tar_interactive = TRUE}
tar_stan_mcmc_rep_summary(
name = mcmc,
stan_files = "model.stan",
data = simulate_data(), # Defined in another code chunk.
batches = tar_toggle(1, 100),
reps = tar_toggle(1, 10),
chains = tar_toggle(1, 4),
parallel_chains = tar_toggle(1, 4),
iter_warmup = tar_toggle(100, 4e4),
iter_sampling = tar_toggle(100, 4e4),
summaries = list(
~posterior::quantile2(.x, probs = c(0.025, 0.25, 0.5, 0.75, 0.975)),
rhat = ~posterior::rhat(.x)
),
deployment = "worker"
)
```C.13 Chunk options
tar_globals: Logical of length 1, whether to define globals or targets. IfTRUE, the chunk code defines functions, objects, and options common to all the targets. IfFALSEorNULL(default), then the chunk returns formal targets for the pipeline.tar_interactive: Logical of length 1 to choose whether to run the chunk in interactive mode or non-interactive mode.tar_name: name to use for writing helper script files (e.g. _targets_r/targets/target_script.R) and specifying target names if the tar_simple chunk option is TRUE. All helper scripts and target names must have unique names, so please do not set this option globally with knitr::opts_chunk$set().tar_script: Character of length 1, where to write the target script file in non-interactive mode. Most users can skip this option and stick with the default_targets.Rscript path. Helper script files are always written next to the target script in a folder with an"_r"suffix. Thetar_scriptpath must either be absolute or be relative to the project root (where you calltar_make()or similar). If not specified, the target script path defaults totar_config_get("script")(default:_targets.R; helpers default:_targets_r/). When you runtar_make()etc. with a non-default target script, you must select the correct target script file either with thescriptargument or withtar_config_set(script = ...). The function willsource()the script file from the current working directory (i.e. withchdir = FALSEinsource()).tar_simple: Logical of length 1. Set toTRUEto define a single target with a simplified interface. In code chunks withtar_simpleequal toTRUE, the chunk label (or thetar_namechunk option if you set it) becomes the name, and the chunk code becomes the command. In other words, a code chunk with labeltargetnameand commandmycommand()automatically gets converted totar_target(name = targetname, command = mycommand()). All other arguments oftar_target()remain at their default values (configurable withtar_option_set()in atar_globals = TRUEchunk).
Target Markdown is powered entirely by
targetsandknitr. It does not actually require Markdown, although Markdown is the recommended way to interact with it.↩︎In
targetsversion 0.6.0, the mode is interactive ifinteractive()isTRUE. In subsequent versions, the mode is interactive if!isTRUE(getOption("knitr.in.progress"))isTRUE.↩︎The
_targets.Rfile from Target Markdown never changes from chunk to chunk or report to report, so you can spread your work over multiple reports without worrying about aligning_targets.Rscripts. Just be sure all your chunk names are unique across all the reports of a project, or you set thetar_namechunk option to specify base names of script file paths.↩︎In addition, for
bookdownprojects, chunk labels should only use alphanumeric characters and dashes.↩︎