Chapter 9 Customer churn and deep learning
drake is designed for workflows with long runtimes, and a major use case is deep learning. This chapter demonstrates how to leverage drake to manage a deep learning workflow. The original example comes from a blog post by Matt Dancho, and the chapter’s content itself comes directly from this R notebook, part of an RStudio Solutions Engineering example demonstrating TensorFlow in R. The notebook is modified and redistributed under the terms of the Apache 2.0 license, copyright RStudio (details here).
9.1 Churn packages
First, we load our packages into a fresh R session.
library(drake)
library(keras)
library(tidyverse)
library(rsample)
library(recipes)
library(yardstick)9.2 Churn functions
drake is R-focused and function-oriented. We create functions to preprocess the data,
prepare_recipe <- function(data) {
data %>%
training() %>%
recipe(Churn ~ .) %>%
step_rm(customerID) %>%
step_naomit(all_outcomes(), all_predictors()) %>%
step_discretize(tenure, options = list(cuts = 6)) %>%
step_log(TotalCharges) %>%
step_mutate(Churn = ifelse(Churn == "Yes", 1, 0)) %>%
step_dummy(all_nominal(), -all_outcomes()) %>%
step_center(all_predictors(), -all_outcomes()) %>%
step_scale(all_predictors(), -all_outcomes()) %>%
prep()
}define a keras model, exposing arguments to set the dimensionality and activation functions of the layers,
define_model <- function(rec, units1, units2, act1, act2, act3) {
input_shape <- ncol(
juice(rec, all_predictors(), composition = "matrix")
)
keras_model_sequential() %>%
layer_dense(
units = units1,
kernel_initializer = "uniform",
activation = act1,
input_shape = input_shape
) %>%
layer_dropout(rate = 0.1) %>%
layer_dense(
units = units2,
kernel_initializer = "uniform",
activation = act2
) %>%
layer_dropout(rate = 0.1) %>%
layer_dense(
units = 1,
kernel_initializer = "uniform",
activation = act3
)
}train a model,
train_model <- function(
rec,
units1 = 16,
units2 = 16,
act1 = "relu",
act2 = "relu",
act3 = "sigmoid"
) {
model <- define_model(
rec = rec,
units1 = units1,
units2 = units2,
act1 = act1,
act2 = act2,
act3 = act3
)
compile(
model,
optimizer = "adam",
loss = "binary_crossentropy",
metrics = c("accuracy")
)
x_train_tbl <- juice(
rec,
all_predictors(),
composition = "matrix"
)
y_train_vec <- juice(rec, all_outcomes()) %>%
pull()
fit(
object = model,
x = x_train_tbl,
y = y_train_vec,
batch_size = 32,
epochs = 32,
validation_split = 0.3,
verbose = 0
)
model
}compare predictions against reality,
confusion_matrix <- function(data, rec, model) {
testing_data <- bake(rec, testing(data))
x_test_tbl <- testing_data %>%
select(-Churn) %>%
as.matrix()
y_test_vec <- testing_data %>%
select(Churn) %>%
pull()
yhat_keras_class_vec <- model %>%
predict_classes(x_test_tbl) %>%
as.factor() %>%
fct_recode(yes = "1", no = "0")
yhat_keras_prob_vec <-
model %>%
predict_proba(x_test_tbl) %>%
as.vector()
test_truth <- y_test_vec %>%
as.factor() %>%
fct_recode(yes = "1", no = "0")
estimates_keras_tbl <- tibble(
truth = test_truth,
estimate = yhat_keras_class_vec,
class_prob = yhat_keras_prob_vec
)
estimates_keras_tbl %>%
conf_mat(truth, estimate)
}and compare the performance of multiple models.
compare_models <- function(...) {
name <- match.call()[-1] %>%
as.character()
df <- map_df(list(...), summary) %>%
filter(.metric %in% c("accuracy", "sens", "spec")) %>%
mutate(name = rep(name, each = n() / length(name))) %>%
rename(metric = .metric, estimate = .estimate)
ggplot(df) +
geom_line(aes(x = metric, y = estimate, color = name, group = name)) +
theme_gray(24)
}9.3 Churn plan
Next, we define our workflow in a drake plan. We will prepare the data, train different models with different activation functions, and compare the models in terms of performance.
activations <- c("relu", "sigmoid")
plan <- drake_plan(
data = read_csv(file_in("customer_churn.csv"), col_types = cols()) %>%
initial_split(prop = 0.3),
rec = prepare_recipe(data),
model = target(
train_model(rec, act1 = act),
format = "keras", # Supported in drake > 7.5.2 to store models properly.
transform = map(act = !!activations)
),
conf = target(
confusion_matrix(data, rec, model),
transform = map(model, .id = act)
),
metrics = target(
compare_models(conf),
transform = combine(conf)
)
)The plan is a data frame with the steps we are going to do.
plan
#> # A tibble: 7 x 3
#> target command format
#> <chr> <expr_lst> <chr>
#> 1 conf_relu confusion_matrix(data, rec, model_relu) … <NA>
#> 2 conf_sigmoid confusion_matrix(data, rec, model_sigmoid) … <NA>
#> 3 data read_csv(file_in("customer_churn.csv"), col_types = cols(… <NA>
#> 4 metrics compare_models(conf_relu, conf_sigmoid) … <NA>
#> 5 model_relu train_model(rec, act1 = "relu") … keras
#> 6 model_sigmo… train_model(rec, act1 = "sigmoid") … keras
#> 7 rec prepare_recipe(data) … <NA>9.4 Churn dependency graph
The graph visualizes the dependency relationships among the steps of the workflow.
vis_drake_graph(plan)9.5 Run the Keras models
Call make() to actually run the workflow.
make(plan)
#> ▶ target data
#> ▶ target rec
#> ▶ target model_relu
#> ▶ target model_sigmoid
#> ▶ target conf_relu
#> ▶ target conf_sigmoid
#> ▶ target metrics9.6 Inspect the Keras results
The two models performed about the same.
readd(metrics) # see also loadd()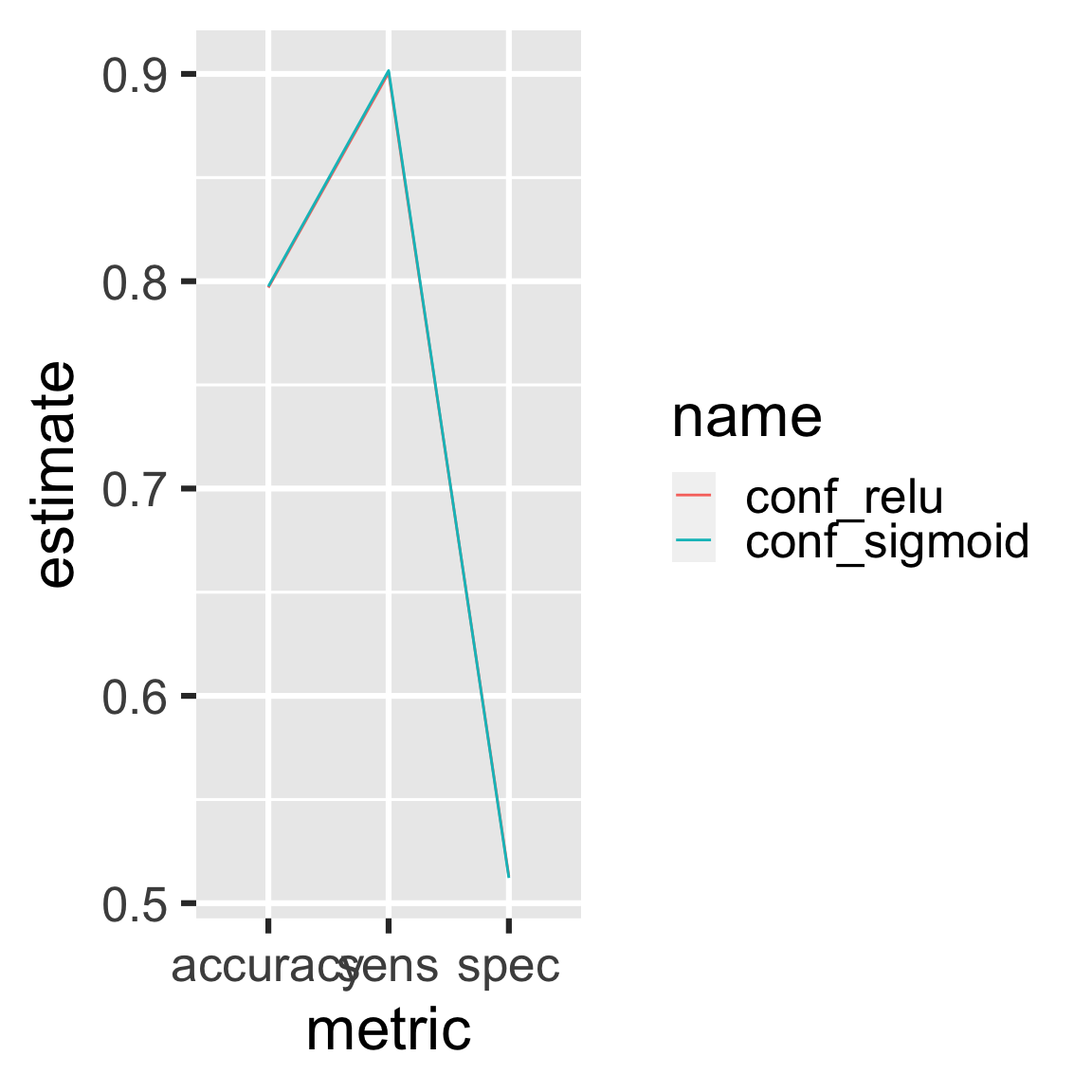
9.7 Add Keras models
Let’s try the softmax activation function.
activations <- c("relu", "sigmoid", "softmax")
plan <- drake_plan(
data = read_csv(file_in("customer_churn.csv"), col_types = cols()) %>%
initial_split(prop = 0.3),
rec = prepare_recipe(data),
model = target(
train_model(rec, act1 = act),
format = "keras", # Supported in drake > 7.5.2 to store models properly.
transform = map(act = !!activations)
),
conf = target(
confusion_matrix(data, rec, model),
transform = map(model, .id = act)
),
metrics = target(
compare_models(conf),
transform = combine(conf)
)
)vis_drake_graph(plan) # see also outdated() and predict_runtime()make() skips the relu and sigmoid models because they are already up to date. (Their dependencies did not change.) Only the softmax model needs to run.
make(plan)
#> ▶ target model_softmax
#> ▶ target conf_softmax
#> ▶ target metrics9.8 Inspect the Churn results again
readd(metrics) # see also loadd()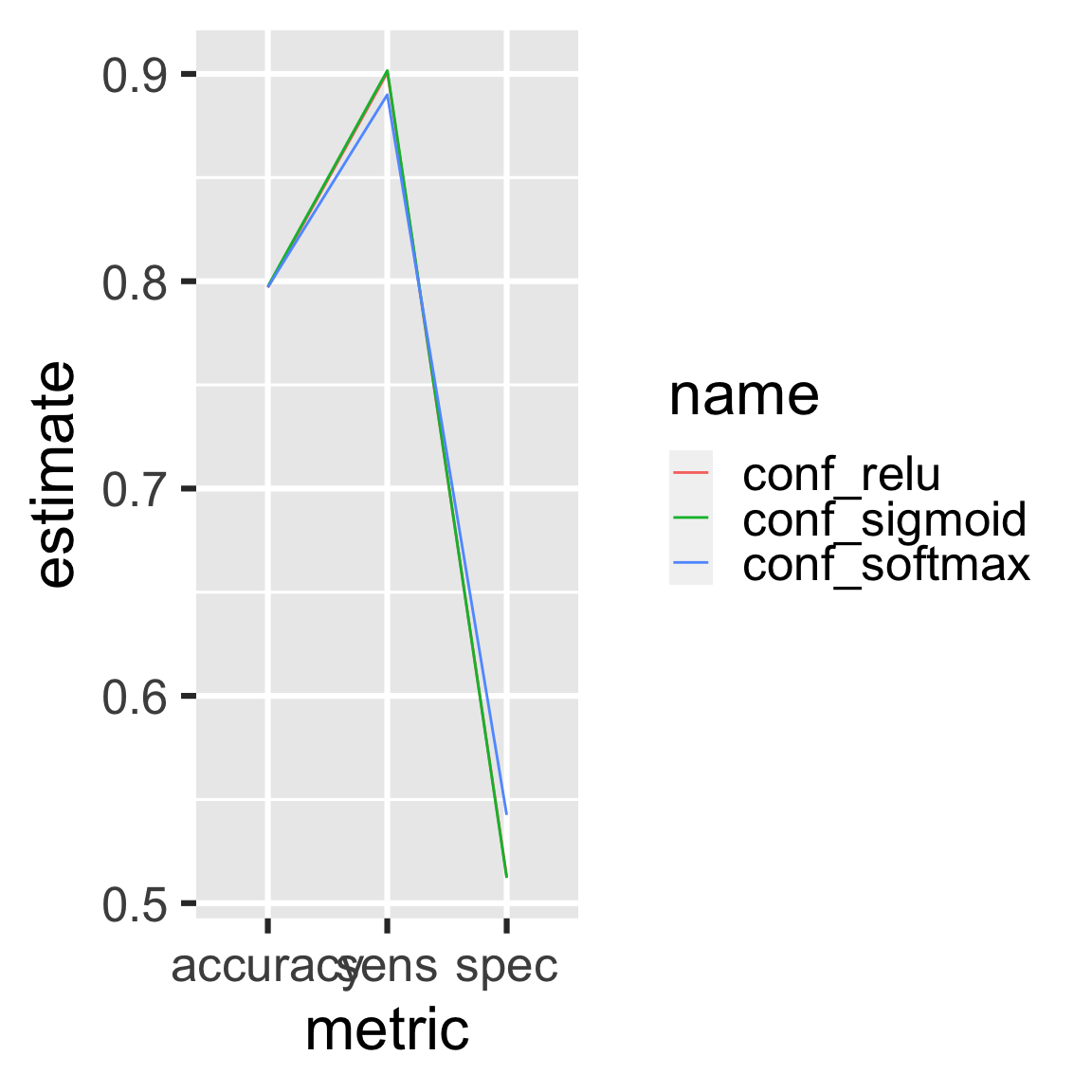
9.9 Update the Churn code
If you change upstream functions, even nested ones, drake automatically refits the affected models. Let’s increase dropout in both layers.
define_model <- function(rec, units1, units2, act1, act2, act3) {
input_shape <- ncol(
juice(rec, all_predictors(), composition = "matrix")
)
keras_model_sequential() %>%
layer_dense(
units = units1,
kernel_initializer = "uniform",
activation = act1,
input_shape = input_shape
) %>%
layer_dropout(rate = 0.15) %>% # Changed from 0.1 to 0.15.
layer_dense(
units = units2,
kernel_initializer = "uniform",
activation = act2
) %>%
layer_dropout(rate = 0.15) %>% # Changed from 0.1 to 0.15.
layer_dense(
units = 1,
kernel_initializer = "uniform",
activation = act3
)
}All the models and downstream results are affected.
make(plan)
#> ▶ target model_relu
#> ▶ target model_sigmoid
#> ▶ target model_softmax
#> ▶ target conf_relu
#> ▶ target conf_sigmoid
#> ▶ target conf_softmax
#> ▶ target metrics9.10 Churn history and provenance
drake tracks history and provenance. You can see which models you ran, when you ran them, how long they took, and which settings you tried (i.e. named arguments to function calls in your commands).
history <- drake_history()
history
#> # A tibble: 17 x 10
#> target current built exists hash command seed runtime prop act1
#> <chr> <lgl> <chr> <lgl> <chr> <chr> <int> <dbl> <dbl> <chr>
#> 1 conf_r… FALSE 2021-02… TRUE 625c… "confusion_… 4.05e8 0.455 NA <NA>
#> 2 conf_r… TRUE 2021-02… TRUE 0e43… "confusion_… 4.05e8 0.385 NA <NA>
#> 3 conf_s… FALSE 2021-02… TRUE 6d39… "confusion_… 1.93e9 0.39 NA <NA>
#> 4 conf_s… TRUE 2021-02… TRUE edd4… "confusion_… 1.93e9 0.344 NA <NA>
#> 5 conf_s… FALSE 2021-02… TRUE 6813… "confusion_… 1.80e9 0.370 NA <NA>
#> 6 conf_s… TRUE 2021-02… TRUE 5079… "confusion_… 1.80e9 0.351 NA <NA>
#> 7 data TRUE 2021-02… TRUE 62d3… "read_csv(f… 1.29e9 0.085 0.3 <NA>
#> 8 metrics FALSE 2021-02… TRUE 00c6… "compare_mo… 1.21e9 0.053 NA <NA>
#> 9 metrics FALSE 2021-02… TRUE d3e7… "compare_mo… 1.21e9 0.075 NA <NA>
#> 10 metrics TRUE 2021-02… TRUE 3a8e… "compare_mo… 1.21e9 0.061 NA <NA>
#> 11 model_… FALSE 2021-02… TRUE a52a… "train_mode… 1.47e9 10.2 NA relu
#> 12 model_… TRUE 2021-02… TRUE 056b… "train_mode… 1.47e9 4.99 NA relu
#> 13 model_… FALSE 2021-02… TRUE 8b5b… "train_mode… 1.26e9 4.93 NA sigm…
#> 14 model_… TRUE 2021-02… TRUE 36e7… "train_mode… 1.26e9 5.02 NA sigm…
#> 15 model_… FALSE 2021-02… TRUE 220e… "train_mode… 8.05e8 5.00 NA soft…
#> 16 model_… TRUE 2021-02… TRUE 4e0c… "train_mode… 8.05e8 5.04 NA soft…
#> 17 rec TRUE 2021-02… TRUE 71f6… "prepare_re… 6.29e8 0.298 NA <NA>And as long as you did not run clean(garbage_collection = TRUE), you can get the old data back. Let’s find the oldest run of the relu model.
hash <- history %>%
filter(act1 == "relu") %>%
pull(hash) %>%
head(n = 1)
drake_cache()$get_value(hash)
#> Model
#> Model: "sequential"
#> ________________________________________________________________________________
#> Layer (type) Output Shape Param #
#> ================================================================================
#> dense_2 (Dense) (None, 16) 576
#> ________________________________________________________________________________
#> dropout_1 (Dropout) (None, 16) 0
#> ________________________________________________________________________________
#> dense_1 (Dense) (None, 16) 272
#> ________________________________________________________________________________
#> dropout (Dropout) (None, 16) 0
#> ________________________________________________________________________________
#> dense (Dense) (None, 1) 17
#> ================================================================================
#> Total params: 865
#> Trainable params: 865
#> Non-trainable params: 0
#> ________________________________________________________________________________